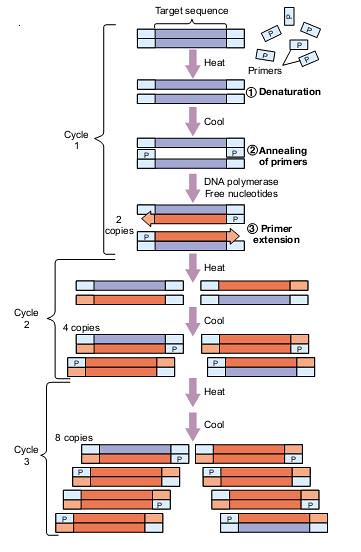
This is a method to
produce very large numbers of copies of specific DNA sequences without cloning.
Therefore PCR can
amplify specific sequences or add sequences such as endonuclease
recognition sequences as primers
to cloned DNA.
PCR consists of 5 main
components.
-Target DNA
-Single stranded
Oligonucleotides (primers)
-d NTPs (dATP, dCTP,
dTTP, dGTP)
-Taq DNA polymerase
-Termocycler
There are three main
steps in PCR.
Step1: Denaturation. The mixture of excess primer and DNA fragment
is heated to about 95° C causing the double strands of target DNA to be denatured
into single strands.
Step 2: Annealing
of Primers. The temperature is
dropped down between about 350- 65°C
As the temperature
decreases the single strands of DNA reassociate into double strands. Large excess
of primer allows two primers anneal or bind to their complementary sequences on
the target DNA leaving the rest of the fragment single-stranded.
Step 3: Primer
Extension. The temperature is
raised to 700- 75°C Taq
polymerase is added. Taq polymerase extends the primer into a
complementary copy of the entire single-stranded fragment, in the 5’-3’
direction. As both the DNA strands are replicated,
two copies of the original fragment are gained.
This process is
repeated many times. At each time, the number of DNA copies doubles. This is
continued until enough copies are gained for the analysis.